Home page Description:
Study uncovers how RNA molecules coded from regions between genes drive prostate cancer.
Posted On: November 09, 2016
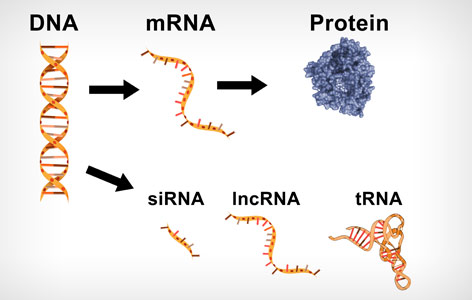
Image Caption:
DNA serves as a template to generate various types of RNA (depicted). Regions of DNA known as genes are copied into mRNA and used by the cell to make proteins.
The basic building blocks of life are proteins. Cells make proteins by reading genetic information (ie, genes) to create a molecule known as RNA—a ‘working copy’ of the genetic code.
Within the cell there are different types of RNA. The most heavily studied type of RNA contains ‘coding genes’ and is used by the cell to make proteins. Another type of RNA, which is ‘non-coding’ and is not used to make proteins, is known as ‘long non-coding RNA’ (lncRNA). This type of RNA has recently been implicated in a number of diseases. Because non-coding genes are less studied, the exact function of lncRNA molecules in the cell is a mystery.
To shed light on lncRNA function, PM Cancer Centre Scientist Dr. Housheng Hansen He carried out a detailed analyses of the role of lncRNA in prostate cancer. Drawing on large, publicly available genetic and genomic datasets, his team identified 45 lncRNAs that may be associated with an increased risk of prostate cancer. The top hit, known as PCAT1, was studied in-depth.
Using experimental models, the researchers found that PCAT1 promotes tumour growth. Furthermore, PCAT1 was found to be involved in cell changes that are linked to prostate cancer development and progression (eg, changes in gene regulation that occur after prolonged exposure to male sex hormones).
“Speckled throughout the genome are discrete sites that have been flagged as associated with cancer risk. While only a fraction of these regions are near protein-coding genes, many more are in close proximity to regions that code for lncRNAs. Exploration of these regions, which are relatively unstudied because they were previously believed to be ‘junk’ DNA, could unveil new targetable drivers of cancer growth,” says Dr. He.
This work was supported by the Canada Foundation for Innovation, the Ontario Research Fund, the Natural Sciences and Engineering Research Council, the Canadian Cancer Society, Canadian Institutes of Health Research, Terry Fox Research Institute, Prostate Cancer Canada, National Institutes of Health and The Princess Margaret Cancer Foundation.
Modulation of long noncoding RNAs by risk SNPs underlying genetic predispositions to prostate cancer. Guo H, Ahmed M, Zhang F, Yao CQ, Li S, Liang Y, Hua J, Soares F, Sun Y,Langstein J, Li Y, Poon C, Bailey SD, Desai K, Fei T, Li Q, Sendorek DH, FraserM, Prensner JR, Pugh TJ, Pomerantz M, Bristow RG, Lupien M, Feng FY, Boutros PC, Freedman ML, Walsh MJ, He HH. Nature Genetics. doi: 10.1038/ng.3637. 2016 Aug 15. [Pubmed abstract]
Within the cell there are different types of RNA. The most heavily studied type of RNA contains ‘coding genes’ and is used by the cell to make proteins. Another type of RNA, which is ‘non-coding’ and is not used to make proteins, is known as ‘long non-coding RNA’ (lncRNA). This type of RNA has recently been implicated in a number of diseases. Because non-coding genes are less studied, the exact function of lncRNA molecules in the cell is a mystery.
To shed light on lncRNA function, PM Cancer Centre Scientist Dr. Housheng Hansen He carried out a detailed analyses of the role of lncRNA in prostate cancer. Drawing on large, publicly available genetic and genomic datasets, his team identified 45 lncRNAs that may be associated with an increased risk of prostate cancer. The top hit, known as PCAT1, was studied in-depth.
Using experimental models, the researchers found that PCAT1 promotes tumour growth. Furthermore, PCAT1 was found to be involved in cell changes that are linked to prostate cancer development and progression (eg, changes in gene regulation that occur after prolonged exposure to male sex hormones).
“Speckled throughout the genome are discrete sites that have been flagged as associated with cancer risk. While only a fraction of these regions are near protein-coding genes, many more are in close proximity to regions that code for lncRNAs. Exploration of these regions, which are relatively unstudied because they were previously believed to be ‘junk’ DNA, could unveil new targetable drivers of cancer growth,” says Dr. He.
This work was supported by the Canada Foundation for Innovation, the Ontario Research Fund, the Natural Sciences and Engineering Research Council, the Canadian Cancer Society, Canadian Institutes of Health Research, Terry Fox Research Institute, Prostate Cancer Canada, National Institutes of Health and The Princess Margaret Cancer Foundation.
Modulation of long noncoding RNAs by risk SNPs underlying genetic predispositions to prostate cancer. Guo H, Ahmed M, Zhang F, Yao CQ, Li S, Liang Y, Hua J, Soares F, Sun Y,Langstein J, Li Y, Poon C, Bailey SD, Desai K, Fei T, Li Q, Sendorek DH, FraserM, Prensner JR, Pugh TJ, Pomerantz M, Bristow RG, Lupien M, Feng FY, Boutros PC, Freedman ML, Walsh MJ, He HH. Nature Genetics. doi: 10.1038/ng.3637. 2016 Aug 15. [Pubmed abstract]




