
Knocking on the door to the world of proteins
When Cheryl Arrowsmith was in university, it did not dawn on her that she would one day become a leader in the field of structural genomics and be involved in collecting and studying a treasure trove of human proteins.
Cheryl studied chemistry during her undergraduate years, and subsequently pursued a PhD in physical organic chemistry at the University of Toronto. “Chemistry clicks with me. I enjoy perceiving everything in terms of atoms and molecules,” says Cheryl.
During her PhD, Cheryl shifted her interests from small molecules to proteins after she took a course in Nuclear Magnetic Resonance (NMR) spectroscopy—a well-established method for analyzing protein structures. Her enjoyment and application of NMR motivated her to continue as a postdoctoral researcher in the field.
Cheryl completed her postdoctoral studies at Stanford’s Magnetic Resonance Lab, where she was supervised by NMR pioneer Dr. Oleg Jardetzky. During this time, she studied how proteins control DNA and gene expression—processes that are key to cancer development.
Her expertise in DNA and protein interactions led her to a career in cancer biology. In 1992, she returned to Toronto to start her own research program at the Princess Margaret Cancer Centre (PM).
Since then, Cheryl has spent the last three decades exploring uncharted territory within structural genomics—through which she has shed new light on cancer biology and therapeutic strategies.
Defining structural genomics on the crest of a technological wave
In the first decade of her research, Cheryl streamlined methods to stably express recombinant proteins in cells and characterized important cancer-related proteins, such as the p53 tumor suppressor.
Cheryl collaborated with Dr. Sam Benchimol, a leader in the p53 field and a former PM scientist, to decode the structure of p53—the most commonly mutated protein in human cancers. As a transcription factor, p53 recognizes and binds to a specific sequence of DNA. Cheryl’s work has demonstrated how p53 forms a tetramer when it binds to DNA to turn gene expression on or off.
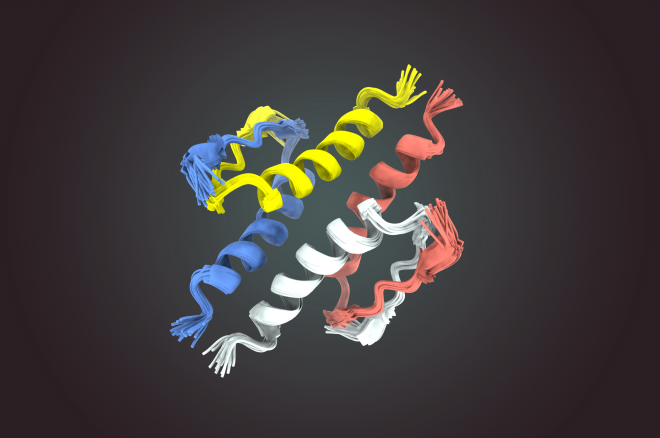
Tetramerization of p53: Four p53 molecules come together to form an active protein complex
“I worked on p53 for about 15 years. Every couple of years, new insights into p53 are uncovered,” says Cheryl. “However, progress was still too slow for just one protein, and there were so many other genes and proteins to study using new and emerging sequencing technologies.”
Motivated by the need to accelerate discovery and to tackle a whole proteome—i.e., the complete set of proteins in an organism—Cheryl’s next project at PM was to uncover the proteome expressed by an archaeon, Methanobacterium thermoautotrophicum. Before scientists unravelled the entire human genome, single-celled microorganisms like bacteria and archaea, were the first entire genomes to be sequenced.
In collaboration with Dr. Aled Edwards, the team published their first experimental paper in the new field of structural genomics. This study defined a systematic workflow on how to conduct large scale protein structure analysis. The study provided a roadmap that led from protein expression and purification, to structural analysis using NMR spectroscopy. This approach was later extended to the method of X-ray crystallography and has become a workhorse for reliably defining protein structures en masse.
In the second decade of her research, following the flood of DNA sequence data from the Human Genome Project (led by the National Institutes of Health) in 2003, Cheryl and Aled seized the opportunity to focus on human proteins.
“A whole new area opened up, which marked the beginning of an exciting decade for structural genomics. The goal was to decipher all human proteins in a fast or even semi-automated fashion, instead of one protein at a time,” recalls Cheryl.
“One of the reasons so many drugs fail in the clinic is because we do not have a sufficient understanding of human biology and disease at the molecular level to be able to reliably know which proteins are good drug targets,” Cheryl explains. “When it was becoming clear that sequencing of the human genome would be completed, pharmaceutical companies pooled their funding together and sought an academic group to work on solving human proteins and finding potential drug targets.”
Built on Cheryl and Aled’s early structural genomic work, and with funding from industry, the Wellcome Trust, and Canadian funders, the Structural Genomic Consortium (SGC) was launched. The initiative is focused on studying proteins that have biomedical relevance and significance to human health. The consortium opened two locations at the onset—one in Oxford, UK and the other in Toronto, Canada. Cheryl took on the role of Chief Scientist and lead of the Toronto site. The team was highly productive—they ambitiously aimed to solve 200 new three-dimensional protein structures a year and they achieved it with unprecedented speed. To date, Cheryl is a co-author on more than 2000 protein structures, which are all available in the Protein DataBank (PDB).
In the current decade, Cheryl’s research focus has transitioned from structural biology to chemical biology, using human proteins and knowledge of 3D structure to make drug-like compounds, called chemical probes, that can modulate the function of proteins.
“These chemical probes can be used as tools to understand a protein’s role in biology and cancer, and they may also be the starting point for further drug development,” says Cheryl.
While computational tools now exist that can predict the rough shape of most proteins with reasonable accuracy, thanks to the massive data in the PDB, it is currently still difficult to predict how molecules, such as drugs, bind to protein targets. Thus, chemical probe discovery still relies on an experimental process that uncovers structures of proteins when they are bound to small chemical compounds.
Under Cheryl’s leadership, the SGC has developed 68 chemical probes as of July 2023. They created a public repository of these probes to facilitate cancer research across the world. “Anybody can use these chemical probes for their research, just like the protein structures in the public protein database.”
According to Cheryl, chemical probes provide more insights than any individual protein structure because they are the link between cell biology and therapeutics. To develop inhibitory compounds for different cancer modalities and to test them in patient-derived disease models, the team collaborates with Drs. Catherine O'Brien (colorectal cancer), Laurie Ailles (clear cell renal carcinoma), Mark Minden (leukemia), Mathieu Lupien, Linda Penn, Benjamin Haibe-Kains and David Cescon(breast cancer) at Princess Margaret; and Dr. Peter Dirks (glioblastoma) at SickKids. The team also works with proteomics expert Dr. Brian Raught and epigenetics experts at Princess Margaret to decode protein interactions and their impact on gene expression.
“Each individual protein is a puzzle piece in the cell. The future of structural genomics is to try to fit the puzzle pieces together. Understanding protein interactions and how they work together will give us deeper insights into cancer and ways to stop it.”
Cultivating the next generation of scientists
Cheryl considers herself an “applier” of technology. Her career has flourished alongside many transformational scientific advancements. From the development of NMR spectroscopy and X-ray crystallography to genomic revolution and the rise of epigenomics, Cheryl keeps her eyes fixed on emerging technologies.
“My biggest piece of advice for young researchers is to stay abreast of what's going on throughout the rest of the world,” says Cheryl. “Stay at the forefront of the crest of the wave, if you will, whatever that wave is, because that's where you can make the most impact.”
Cheryl’s commitment to staying at the cutting edge has positioned her as a highly influential leader and mentor. She was recently awarded the 2023 Richard Hill Mentorship Award at the Princess Margaret Annual Research Retreat. Over the past two decades, Cheryl has provided direct mentorship to a significant number of individuals, including 41 postdoctoral fellows, 15 graduate students, over 20 undergraduate students and more than 30 other highly qualified personnel. Her mentorship has consistently attracted exceptional trainees, with ten of her former research mentees securing faculty positions globally.
In addition to her scientific endeavors, she finds inspiration in her interests outside of the lab—gardening and beekeeping. Just as she nurtures and cultivates the growth of her plants and bees, Cheryl applies the same care and dedication to mentoring individuals, and fostering their personal and professional development.
“We have made a lot of progress, and I look forward to an even brighter future—one in which we make even greater strides towards advancing drug discovery and the development of much needed medicines,” Cheryl adds.
Meet PMResearch is a monthly column that features Princess Margaret researchers. It showcases the research of world class scientists, as well as their passions and interests in career and life—from hobbies and avocations to career trajectories and life philosophies. The researchers that we select are relevant to advocacy/awareness initiatives, or have recently received awards or published papers. We are also showcasing the diversity of our staff in keeping with UHN themes and priorities.




